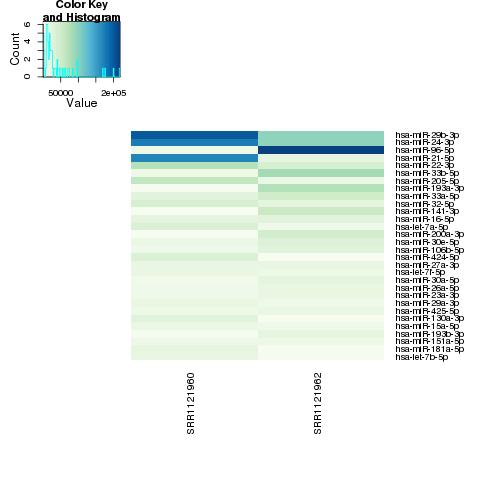
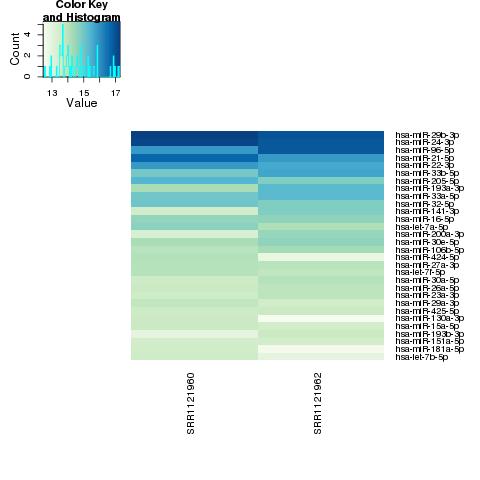
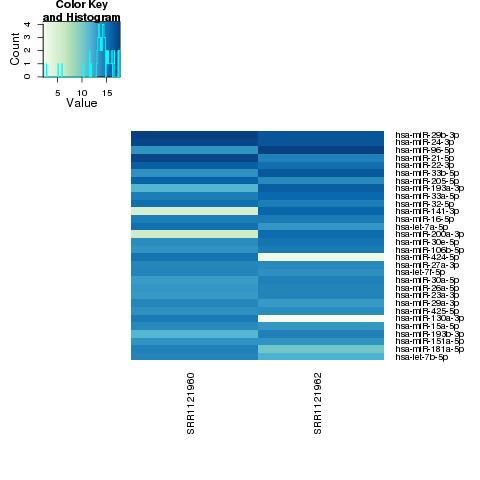
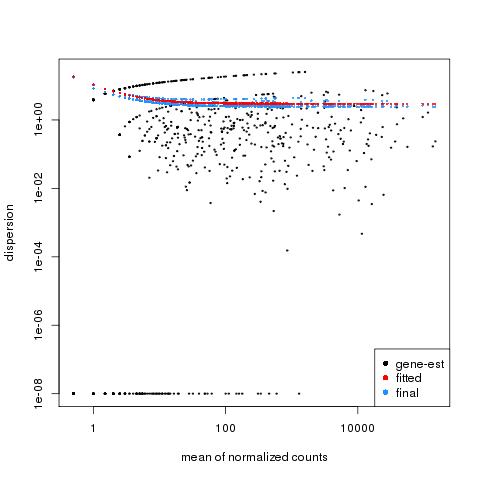
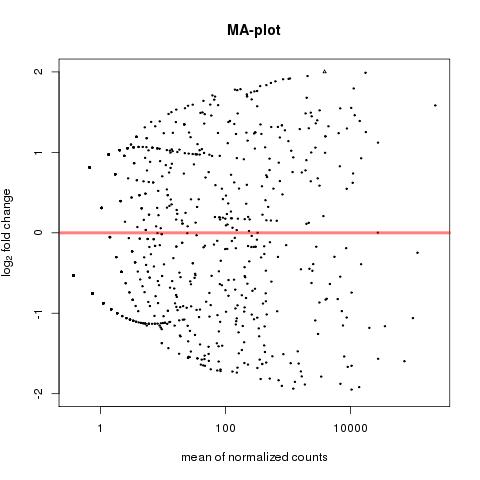
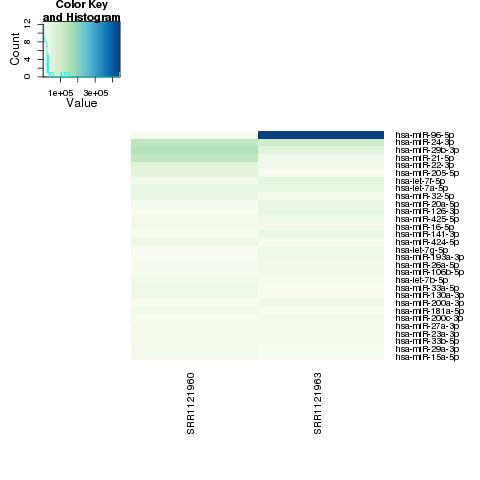
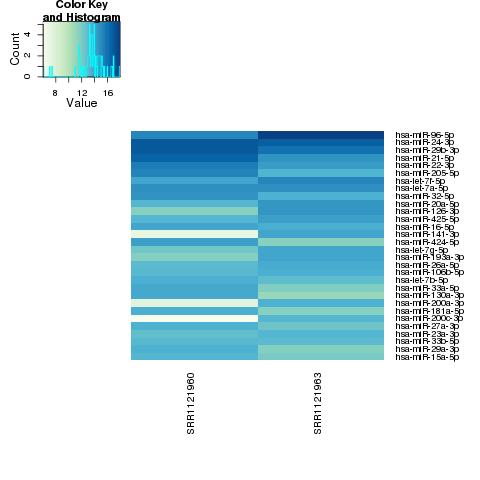
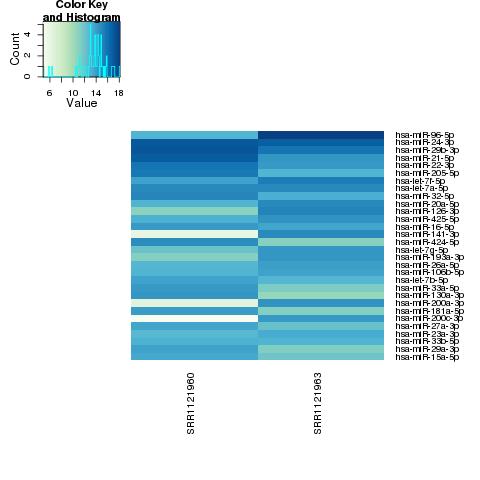
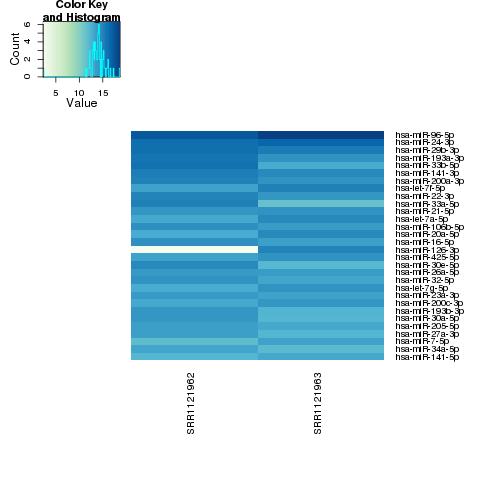
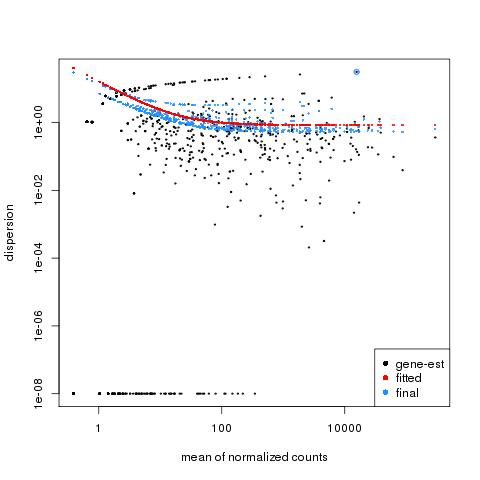
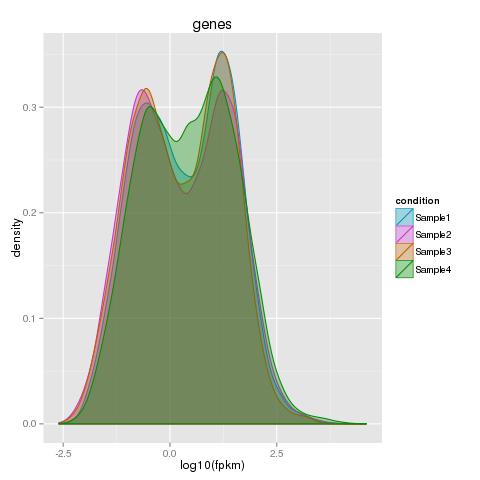
| 49 +/- 40% |
- |
47 |
46 |
0 |
1 |
yes |
- |
cucuccgccaccuccaccgcgg |
gcggcggaggcggcggug |
gcggcggaggcggcgguggcggcagcgguuguuccugccucuccgccaccuccaccgcgg |
2:88355217..88355277:- |
| 49 +/- 40% |
- |
16 |
14 |
0 |
2 |
yes |
- |
cgccgcuuucugggcucgcuca |
ugcgggcccugggagugga |
ugcgggcccugggaguggagacgcggagcugcggucgccgcuuucugggcucgcuca |
5:108084335..108084392:- |
| 59 +/- 35% |
- |
14 |
10 |
0 |
4 |
yes |
- |
agaucaaaagccucagggcaga |
ugcccugagacuuuugcucuaa |
ugcccugagacuuuugcucuaauaauuuauucuaauaauaauuuagaucaaaagccucagggcaga |
3:127305953..127306019:- |
| 74 +/- 22% |
- |
12 |
8 |
0 |
4 |
yes |
- |
uauauauguuguguauguau |
auauacacauauacauauauauguu |
auauacacauauacauauauauguugugaauauucuaagcauauauauguuguguauguau |
11:44254691..44254752:+ |
| 74 +/- 22% |
- |
12 |
10 |
0 |
2 |
yes |
- |
aaauguaauugcuguuuuugcc |
caaaaacagcaauuacuuuugc |
aaauguaauugcuguuuuugccuuuaguuuuaaugacaaaaacagcaauuacuuuugc |
2:78484139..78484197:- |
| 81 +/- 16% |
- |
9 |
8 |
0 |
1 |
yes |
- |
uccgguaggguucgggccuuccg |
gaagacccgagccugccgggg |
uccgguaggguucgggccuuccggaugcggcuugggcgcucuucggggaccuccguggcgcggaagacccgagccugccgggg |
11:118965340..118965423:- |
| 81 +/- 16% |
- |
9 |
6 |
0 |
3 |
yes |
- |
uguguuuguguauguguaugu |
gcacacauacaaacaaacacaca |
gcacacauacaaacaaacacacacaaaaaacgggcauguguguuuguguauguguaugu |
19:43772416..43772475:+ |
| 85 +/- 11% |
- |
6 |
4 |
1 |
1 |
yes |
- |
ucggccucgcucccgccccucc |
caggggcgggcgcggagggcggg |
caggggcgggcgcggagggcgggggcgcgguccgcuagagcccucggccucgcucccgccccucc |
8:80680197..80680262:- |
| 85 +/- 11% |
- |
7 |
6 |
0 |
1 |
yes |
- |
gagguccgcagcugcucugccu |
acagagcagcuguuggaucccc |
acagagcagcuguuggauccccaccaagugucuggagguccgcagcugcucugccu |
14:23447393..23447449:- |
| 85 +/- 11% |
- |
6 |
5 |
0 |
1 |
yes |
- |
cuagaauggauuuuuacaugcc |
cauguaaaaauccauucuagc |
cauguaaaaauccauucuagccuugauucuccuguaggcuagaauggauuuuuacaugcc |
2:156603708..156603768:- |
| 87 +/- 10% |
- |
5 |
2 |
0 |
3 |
yes |
- |
ugagggagcaguggcuggggu |
ucucagccucuauucccuggcug |
ucucagccucuauucccuggcuguccccuuuguuugaagcuccagugagggagcaguggcuggggu |
16:57673361..57673427:+ |
| 87 +/- 10% |
- |
5 |
3 |
0 |
2 |
yes |
- |
uaucucauuuggaucuuaaaug |
uuuaggauccaaaugagauaua |
uuuaggauccaaaugagauauaagucuacggguaugucucuuuaucucauuuggaucuuaaaug |
5:63936000..63936064:- |
| 89 +/- 8% |
- |
4 |
3 |
0 |
1 |
yes |
- |
ccuccucgucggccucuucggc |
gggaagauggcggccggc |
gggaagauggcggccggcggcggcggaggcagcaguaaggccuccuccucgucggccucuucggc |
1:150337185..150337250:+ |
| 89 +/- 8% |
- |
13 |
12 |
0 |
1 |
no |
- |
uuugggauugacgccacaugucu |
ucaagugucaucugucccuag |
uuugggauugacgccacaugucucagguccccagcugagucaagugucaucugucccuag |
2:219144849..219144909:- |
| 89 +/- 8% |
- |
45 |
45 |
0 |
0 |
yes |
- |
ugucccggcccggagccgcc |
cggcacccggccugggacgcg |
cggcacccggccugggacgcguagaggcgcacgugucccggcccggagccgcc |
17:17739750..17739803:+ |
| 89 +/- 8% |
- |
65 |
65 |
0 |
0 |
yes |
- |
uauccuccaguagacuagggag |
ccccagccuacuggaggauaag |
ccccagccuacuggaggauaagaggauauaaaggucucuuauccuccaguagacuagggag |
8:99405893..99405954:- |
| 87 +/- 7% |
- |
10 |
7 |
0 |
3 |
no |
- |
cgcgccugcaggaacugguaga |
uuccagcccugguaggcgccgcg |
uuccagcccugguaggcgccgcggaagaaggggaagcgcgccugcaggaacugguaga |
6:1390558..1390616:- |
| 87 +/- 7% |
- |
3 |
2 |
0 |
1 |
yes |
- |
uucuaggccccaccccaagccu |
aacucugaggaggggcccagaag |
uucuaggccccaccccaagccuacugaaccaggaacucugaggaggggcccagaag |
3:124829445..124829501:- |
| 87 +/- 7% |
- |
20 |
20 |
0 |
0 |
yes |
- |
aggagggcacuggcccca |
gggccucuuugcaugcccucugg |
gggccucuuugcaugcccucugggugccggcucaggagggcacuggcccca |
21:45237173..45237224:+ |
| 87 +/- 7% |
- |
13 |
13 |
0 |
0 |
yes |
- |
cugggcuuguggggaccccugg |
agggauccccagacagccccgagcu |
agggauccccagacagccccgagcucugcuagcugggcuuguggggaccccugg |
14:105147059..105147113:+ |
| 87 +/- 7% |
- |
22 |
22 |
0 |
0 |
yes |
- |
aggcugugaugcucuccugagccc |
gcucggcccccacagcgaaacggccgcc |
gcucggcccccacagcgaaacggccgccuaaaccacccaggccuuauggcuucauaggcugugaugcucuccugagccc |
19:11606358..11606437:- |
| 87 +/- 7% |
- |
69 |
69 |
0 |
0 |
yes |
- |
caccgacucugucuccugcaga |
agcaggggagagagaggaguc |
agcaggggagagagaggaguccucuagacaccgacucugucuccugcaga |
17:39673416..39673466:- |
| 87 +/- 7% |
- |
49 |
49 |
0 |
0 |
yes |
- |
cccuguccuccaggagcuc |
gcaaggcccuggggguggggcc |
gcaaggcccuggggguggggcccuggaggugucccggaaaaaacggaacaccuguccuccaggaacacccuguccuccaggagcuc |
22:32524270..32524356:+ |
| 87 +/- 7% |
- |
5 |
3 |
0 |
2 |
yes |
- |
uaugaaaauuaaaggauuuuugu |
uugaaauccuuuucuuuucaagu |
uaugaaaauuaaaggauuuuuguugaauuacugugaauaguguuuuaaaauugaaauccuuuucuuuucaagu |
10:33212667..33212740:- |
| 87 +/- 7% |
- |
16 |
16 |
0 |
0 |
yes |
- |
ggcuccaccuuccuagguuggc |
cagcuuugcagguggugacuag |
ggcuccaccuuccuagguuggcuuugcucucaugccagcuuugcagguggugacuag |
22:26968292..26968349:- |
| 87 +/- 7% |
- |
21 |
15 |
0 |
6 |
yes |
- |
guugcuuaccagaaguucaaau |
gaauuucagguaaacaaugaau |
gaauuucagguaaacaaugaauaguuuuuuagucugucguugcuuaccagaaguucaaau |
1:160312274..160312334:- |
| 42 +/- 13% |
- |
10 |
10 |
0 |
0 |
yes |
- |
agggcgcaguuccgggcgcguc |
cgcgccaggagcugcgcgggcgcugg |
cgcgccaggagcugcgcgggcgcugggagggagaccagggcgcaguuccgggcgcguc |
17:1733067..1733125:- |
| 42 +/- 13% |
- |
18 |
18 |
0 |
0 |
yes |
- |
gauccaacagcugcucugugagc |
aggcagagcagcugcggaccuc |
aggcagagcagcugcggaccuccagacacuugguggggauccaacagcugcucugugagc |
14:23447394..23447454:+ |
| 42 +/- 13% |
- |
51 |
51 |
0 |
0 |
yes |
- |
uauguuuuccugaggagauau |
aucuccacaggaugccauagu |
aucuccacaggaugccauagugaauagugcuauguuuuccugaggagauau |
1:246996753..246996804:- |
| 42 +/- 13% |
- |
61 |
61 |
0 |
0 |
yes |
- |
aauagcucagaaugucaguucug |
gaauugauaacugagcaaggaa |
aauagcucagaaugucaguucuguuuuaaguaacagaauugauaacugagcaaggaa |
8:101715195..101715252:- |
| 42 +/- 13% |
- |
9 |
9 |
0 |
0 |
yes |
- |
gucccggucgccgcgguucgccg |
guccggcgaccggcccac |
gucccggucgccgcgguucgccgccgccccugguggcgguccggcgaccggcccac |
16:33966064..33966120:+ |
| 42 +/- 13% |
- |
53 |
53 |
0 |
0 |
yes |
- |
gugugcucgcuucggcagcacaua |
ugugugugugugugugugugugugu |
uguguguguguguguguguguguguguguguguguguguuaauaaacucauaacgugugcucgcuucggcagcacaua |
16:69312798..69312876:+ |
| 42 +/- 13% |
- |
17 |
12 |
5 |
0 |
yes |
- |
gccuccuuagcgcaguagg |
cguugcucaaggagugaa |
gccuccuuagcgcaguagguagcacgucagucucauaaucugaagauuucaacaagugagugccucguugcucaaggagugaa |
8:125628997..125629080:- |
| 42 +/- 13% |
- |
9 |
9 |
0 |
0 |
yes |
- |
uaucugcuguuguccccucagg |
agaggccgcacagcaggaacc |
agaggccgcacagcaggaaccucacauuggauguaucugcuguuguccccucagg |
19:7533657..7533712:+ |
| 42 +/- 13% |
- |
9 |
9 |
0 |
0 |
yes |
- |
ugaggccgagaaggcaaccgcga |
gcguuguccucgcgccuucgcc |
gcguuguccucgcgccuucgcccuggguaggugaggccgagaaggcaaccgcga |
1:150207085..150207139:+ |
| 42 +/- 13% |
- |
9 |
8 |
0 |
1 |
yes |
- |
augugugacauuuucucauucu |
ugagaaaaugucccacauuugu |
ugagaaaaugucccacauuuguggacauuugcggacaaugugugacauuuucucauucu |
1:214673355..214673414:- |
| 42 +/- 13% |
- |
73 |
73 |
0 |
0 |
yes |
- |
guucuuccuuggaugucugagugag |
gguucagguauucagaaguuug |
guucuuccuuggaugucugagugagcaucuucauucauugugcuugaacacaaugaguugguucagguauucagaaguuug |
11:17096143..17096224:- |
| 0 +/- 1% |
- |
7 |
7 |
0 |
0 |
yes |
- |
cggccuggcggacgcggucggg |
cgccgccuccgccgggcucgcc |
cgccgccuccgccgggcucgccgggaccuccgggcggccuggcggacgcggucggg |
22:40390801..40390857:- |
| 0 +/- 1% |
- |
16 |
16 |
0 |
0 |
yes |
- |
uuggcugaugaauacugacu |
acaguguuuauguguuuucc |
uuggcugaugaauacugacuguauuuuccuugagcaugucuggaacaguguuuauguguuuucc |
1:173834848..173834912:- |
| 0 +/- 1% |
- |
8 |
8 |
0 |
0 |
yes |
- |
ugugagguugucaugccugc |
aggcaugacaaccucauacu |
ugugagguugucaugccugcuauuacuacagcuaugccagcaggcaugacaaccucauacu |
2:133191304..133191365:+ |
| 0 +/- 1% |
- |
8 |
8 |
0 |
0 |
yes |
- |
ucacugggaggcaucuggagcugau |
uggcugcaggccaccaaguugca |
uggcugcaggccaccaaguugcagguucugacagcuccacugucacugggaggcaucuggagcugau |
17:20378978..20379045:- |
| 0 +/- 1% |
- |
8 |
8 |
0 |
0 |
yes |
- |
ucacugggaggcaucuggagcugau |
uggcugcaggccaccaaguugca |
uggcugcaggccaccaaguugcagguucugacagcuccacugucacugggaggcaucuggagcugau |
17:18371900..18371967:+ |
| 0 +/- 1% |
- |
6 |
6 |
0 |
0 |
yes |
- |
uggcuggcccguggggcgag |
cgucccacgggccagccccg |
cgucccacgggccagccccgccguggccguggccguggcuggcccguggggcgag |
2:27294394..27294449:- |
| 0 +/- 1% |
- |
7 |
7 |
0 |
0 |
yes |
- |
ccagggcaggcgcuuguggaa |
ccaccgaugccuuguuccugagcc |
ccaccgaugccuuguuccugagccuuugcugggagucggccagggcaggcgcuuguggaa |
14:105912945..105913005:+ |
| 0 +/- 1% |
- |
10 |
10 |
0 |
0 |
yes |
- |
gguuccauaguguagugg |
aacuucacauuguauccuu |
aacuucacauuguauccuuaacaugguuccauaguguagugg |
12:71892248..71892290:+ |
| 0 +/- 1% |
- |
7 |
7 |
0 |
0 |
yes |
- |
ccagggcaggcgcuuguggaa |
ccacugccuugcccaccgg |
ccagggcaggcgcuuguggaaggaaggugucccuuaaccccacugccuugcccaccgg |
14:105912984..105913042:+ |